Research
Research Topic #1 – How Plant Cells Control Transposable Elements (and Transgenes)
The Slotkin laboratory investigates how plant cells determine which fragments of DNA should be expressed (like genes), and which should be targeted for repression and not allowed to express. The regions of the genome that are targeted for repression are called Transposable Elements (TEs), which are mobile ‘jumping genes’ that cause mutations when they jump. Most of the time these new mutations generated by TEs hurt the cell or are inconsequential, but occasionally something new is produced that is helpful and will be selected for, leading to some spectacular examples of TE-induced gene or trait regulation.
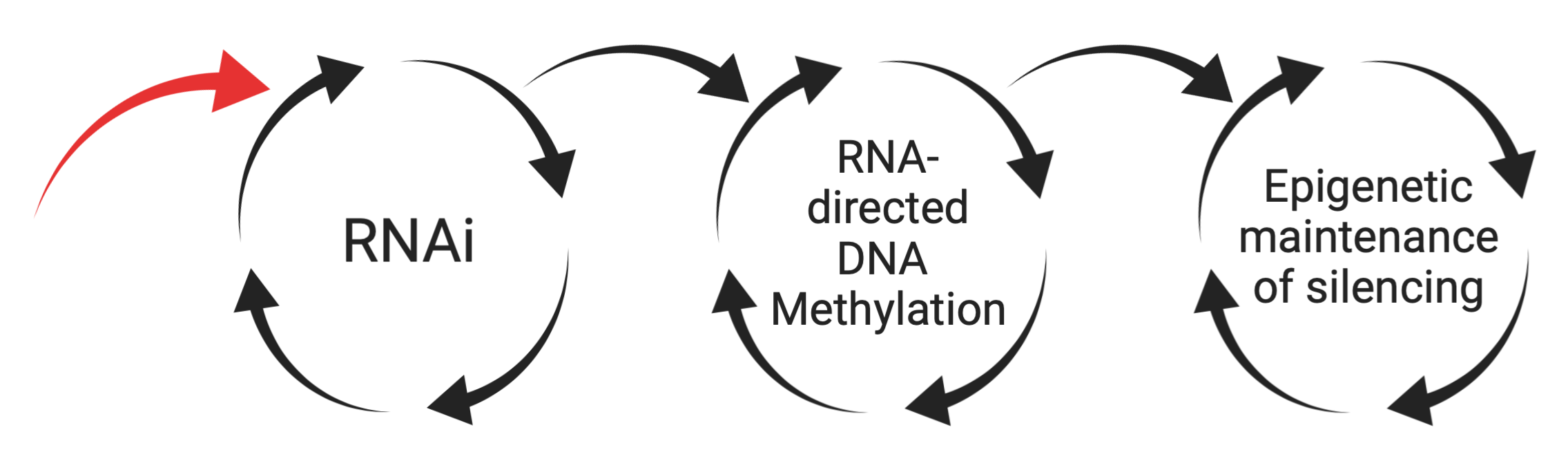
In a broad sense, what the Slotkin laboratory aims to determine is how self-perpetuating feed-forward cycles in the cell are initiated. The cycles we work on are RNA interference (RNAi), RNA-directed DNA Methylation (RdDM) and maintenance epigenetic silencing. We have learned that these cycles can feed into each other. Our goal now is to determine how the first cycle of RNAi is triggered to initiate (red arrow below).
While these three mechanisms evolved to repress transposable elements, they often target the transgenes that researchers add to plant genomes for research or crop improvement. This targeting of transgenes generates unintended and problematic silencing.
If you need help with one of these topics, please feel free to email us and request a collaboration!
Research Topic #2 – How We Can Control Transposable Elements As Tools For Genome Engineering
After more than a decade of elucidating how cells control TEs, we have learned how we can control TE activity. This includes the engineering of the timing of transposition, the location of transposition and the DNA cargo that the TE delivers to the new location. We have combined TEs with gene editing tools such as CRISPR/Cas and Cre/Lox to create new tools to manipulate plant genomes. This is a use-inspired synthetic biology project to create proof-of-principle demonstrations of new technology, and then translate these discoveries into important crops that need new tools. See the details in this preprint.
See our Publications, Press and Resources pages for links to our research articles, while these five Review / Commentary articles below are good representations of our opinions and viewpoints:
Hung, Y.-H., Slotkin, R.K. (2021). The initiation of RNA interference (RNAi) in plants. Current Opinion in Plant Biology 61, 102014.
Shahid, S., Slotkin, R.K. (2020). The current revolution in transposable element biology enabled by long reads. Current Opinion in Plant Biology 54, 49-56.
Slotkin, R.K. (2018). The case for not masking away repetitive DNA. Mobile DNA 9, 475.
Cuerda-Gil, D., Slotkin, R.K. (2016). Non-canonical RNA-directed DNA methylation. Nature Plants 2(11), 16163.
Sigman, M., Slotkin, R.K. (2016). The First Rule of Plant Transposable Element Silencing: Location, Location, Location. The Plant Cell 28(2), 304-313.
Martinez, G., Slotkin, R.K. (2012). Developmental relaxation of transposable element silencing in plants: functional or byproduct? Current Opinion in Plant Biology 15, 496-502.